Set up
If you work on Midway2, some of the steps have been done for you (checked squares)
- install anaconda/miniconda
- define imlabtools conda environment how to here, which will install all the python modules needed for this analysis session
- download data and software from Box. This will have copies of all the software repositories and the models
-
download software (already in )
- download metaxcan repo
- download torus repo
- download fastenloc repo
- download TMWR repo
- download prediction models from predictdb.org (a few models are included in /project2/bios25328/Data/Lab-QGT)
Summary of analysis plan
- predict whole blood expression
- check how well the prediction works with GEUVADIS expression data
- run association between predicted expression and a simulated phenotype
- calculate association between expression levels and coronary artery disease risk using s-predixcan
- fine-map the coronary artery disease gwas results using torus
- calculate colocalization probability using fastenloc
- run transcriptome-wide mendelian randomization in one locus of interest
Preliminary definitions
- log into Midway2 and run the following commands. Don’t forget to replace username with your user name.
# define environmental variables for convenience
USERNAME="WRITE YOUR USER NAME HERE"
PRE="/project2/bios25328/Data/Lab-QGT/"
WORK="/project2/bios25328/username/$USERNAME/lab-QGT/"
SOFTWARE="/project2/bios25328/software/"
## create the working directory if you don't have it
mkdir $WORK
cd $WORK
LAB="/project2/bios25328/Data/Lab-QGT/QGT-Columbia-HKI-repo/"
CODE=$LAB/code
DATA=$PRE/data
MODEL=$PRE/models
RESULTS=$PRE/results
METAXCAN=$PRE/repos/MetaXcan-master/software
TWMR=$PRE/repos/TWMR-master# load python
module load python
# load conda
module load python/anaconda-2020.02
- activate the the imlabtools environment, which will make sure all the necessary python modules are available to the software we will be running.
conda activate $SOFTWARE/MetaXcan/env/imlabtools/Reminder: the bash chunks need to be copy-pasted to the terminal, not performed within the chunk.
- Get some helper scripts
## git clone https://github.com/hakyimlab/QGT-Columbia-HKI-repo/This should have generated a folder /project2/bios25328/Data/Lab-QGT/QGT-Columbia-HKI-repo
- Load and open R
module load R
R- Within R, load the tidyverse package
suppressPackageStartupMessages(library(tidyverse))- define some variables to access the data more easily within the R session. Run the following r chunk
print(getwd())
PRE="/project2/bios25328/Data/Lab-QGT"
MODEL=glue::glue("{PRE}/models")
DATA=glue::glue("{PRE}/data")
RESULTS=glue::glue("{PRE}/results")
METAXCAN=glue::glue("{PRE}/repos/MetaXcan-master/software")
FASTENLOC=glue::glue("{PRE}/repos/fastenloc-master")
TORUS=glue::glue("{PRE}/repos/torus-master")
TWMR=glue::glue("{PRE}/repos/TWMR-master")
lab="/project2/bios25328/Data/Lab-QGT/QGT-Columbia-HKI-repo"
CODE=glue::glue("{lab}/code")
source(glue::glue("{CODE}/load_data_functions.R"))
source(glue::glue("{CODE}/plotting_utils_functions.R"))
# This is a reference table we'll use a lot throughout the lab. It contains information about the genes.
gencode_df = load_gencode_df()Transcriptome-wide association methods
Transcriptome-wide association methods
predict expression
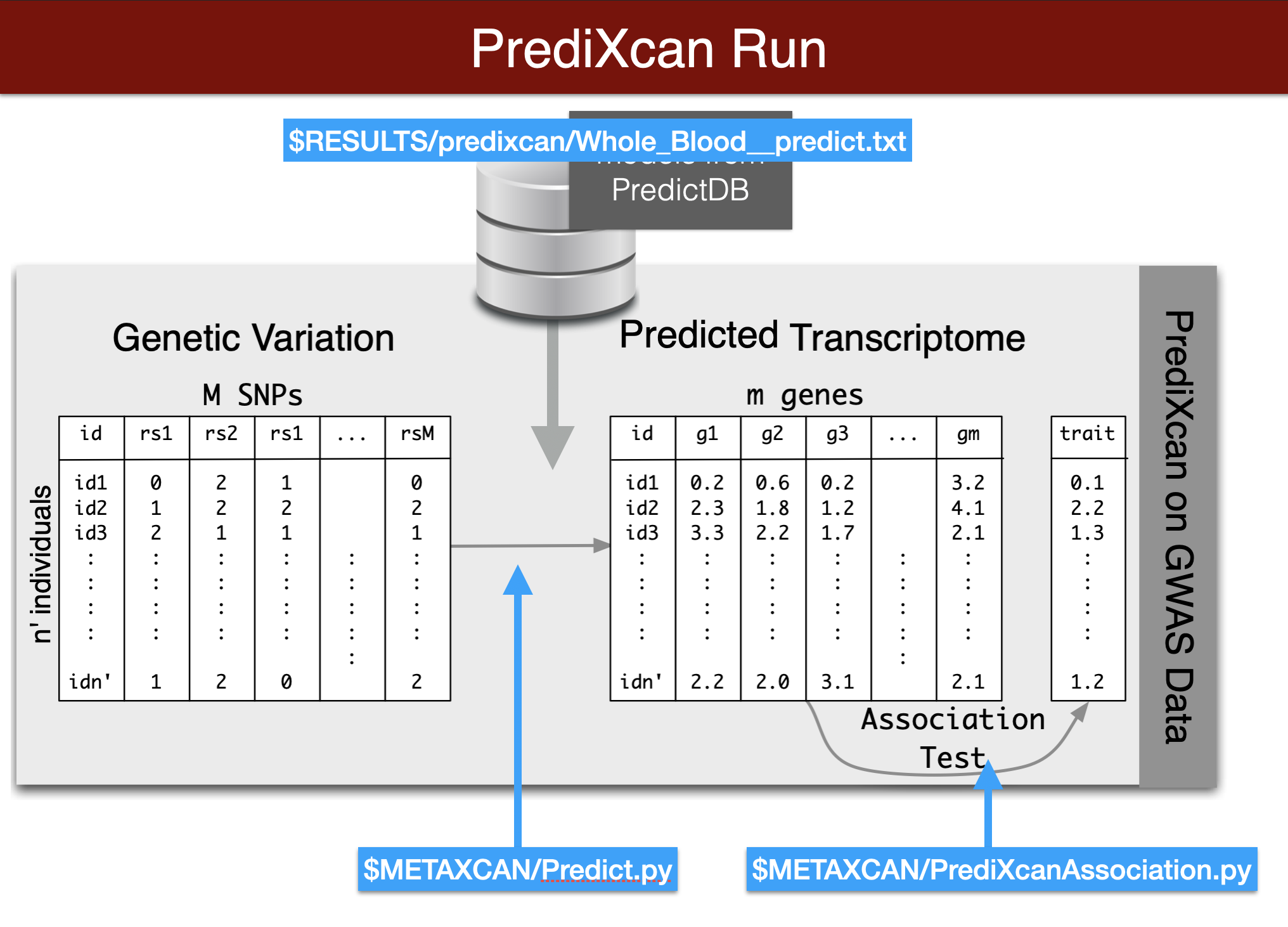
Visual summary of predixcan runs
We will predict expression of genes in whole blood using the Predict.py code in the METAXCAN folder.
Prediction models are located in the MODEL folder. Additional models for different tissues and transcriptome studies can be downloaded from predictdb.org
Remember you need to copy and paste this code chunk into the terminal to run it. Also make sure you activated the imlabtools environment which has all the necessary python modules.
Make sure all the paths and file names are correct
This run should take about one minute.
copy and paste the following block to the terminal and hit enter
printf "Predict expression\n\n"
python3 $METAXCAN/Predict.py \
--model_db_path $PRE/models/gtex_v8_en/en_Whole_Blood.db \
--vcf_genotypes $DATA/predixcan/genotype/filtered.vcf.gz \
--vcf_mode genotyped \
--variant_mapping $DATA/predixcan/gtex_v8_eur_filtered_maf0.01_monoallelic_variants.txt.gz id rsid \
--on_the_fly_mapping METADATA "chr{}_{}_{}_{}_b38" \
--prediction_output $RESULTS/predixcan/Whole_Blood__predict.txt \
--prediction_summary_output $RESULTS/predixcan/Whole_Blood__summary.txt \
--verbosity 9 \
--throw
- check predicted values
prediction_fp = glue::glue("{RESULTS}/predixcan/Whole_Blood__predict.txt")
## Read the Predict.py output into a dataframe. This function reorganizes the data and adds gene names.
predicted_expression = load_predicted_expression(prediction_fp, gencode_df)
head(predicted_expression)
## read summary of prediction, number of SNPs per gene, cross validated prediction performance
prediction_summary = load_prediction_summary(glue::glue("{RESULTS}/predixcan/Whole_Blood__summary.txt"), gencode_df)
## number of genes with a prediction model
dim(prediction_summary)
head(prediction_summary)
print("distribution of prediction performance r2")
summary(prediction_summary$pred_perf_r2)assess prediction performance (optional)
## download and read observed expression data from GEUVADIS
## from https://uchicago.box.com/s/4y7xle5l0pnq9d1fwmthe2ewhogrnlrv
## Remove the version number from the gene_id's (ENSG000XXX.ver)
head(predicted_expression)
## merge predicted expression with observed expression data (by IID and gene)
## plot observes vs predicted expressioni for
## ERAP1 (ENSG00000164307)
## PEX6 (ENSG00000124587)
## calculate spearman correlation for all genes
## what's the best performing gene?run association with a simulated phenotype
\(Y = \sum_k T_k \beta_k + \epsilon\)
with random effects \(\beta_k \sim (1-\pi)\cdot \delta_0 + \pi\cdot N(0,1)\)
sPHENO="sim.spike_n_slab_0.01_pve0.1"
printf "association\n\n"
python3 $METAXCAN/PrediXcanAssociation.py \
--expression_file $RESULTS/predixcan/Whole_Blood__predict.txt \
--input_phenos_file $DATA/predixcan/phenotype/$PHENO.txt \
--input_phenos_column pheno \
--output $RESULTS/predixcan/$PHENO/Whole_Blood__association.txt \
--verbosity 9 \
--throw
More predicted phenotypes can be found in $DATA/predixcan/phenotype/. The naming of the phenotypes provides information about the genic architecture: the number after pve is the proportion of variance of Y explained by the genetic component of expression. The number after spike_n_slab represents the probability that a gene is causal \(\pi\)(i.e. prob \(\beta \ne 0\))
read association results
## read association results
PHENO="sim.spike_n_slab_0.01_pve0.1"
predixcan_association = load_predixcan_association(glue::glue("{RESULTS}/predixcan/{PHENO}/Whole_Blood__association.txt"),
gencode_df)
## take a look at the results
dim(predixcan_association)
predixcan_association %>% arrange(pvalue) %>% head
predixcan_association %>% arrange(pvalue) %>% ggplot(aes(pvalue)) + geom_histogram(bins=20)
## compare distribution against the null (uniform)
gg_qqplot(predixcan_association$pvalue, max_yval = 40)compare estimated effects with true effect sizes
truebetas = load_truebetas(glue::glue("{DATA}/predixcan/phenotype/gene-effects/{PHENO}.txt"), gencode_df)
betas = (predixcan_association %>%
inner_join(truebetas,by=c("gene"="gene_id")) %>%
select(c('estimated_beta'='effect',
'true_beta'='effect_size',
'pvalue',
'gene_id'='gene',
'gene_name'='gene_name.x',
'region_id'='region_id.x')))
betas %>% arrange(pvalue) %>% head
## do you see examples of potential LD contamination?
betas %>% ggplot(aes(estimated_beta, true_beta))+geom_point()+geom_abline()Summary PrediXcan
Now we will use the summary results from a GWAS of coronary artery disease to calculate the association between the genetic component of the expression of genes and coronary artery disease risk. We will use the SPrediXcan.py.
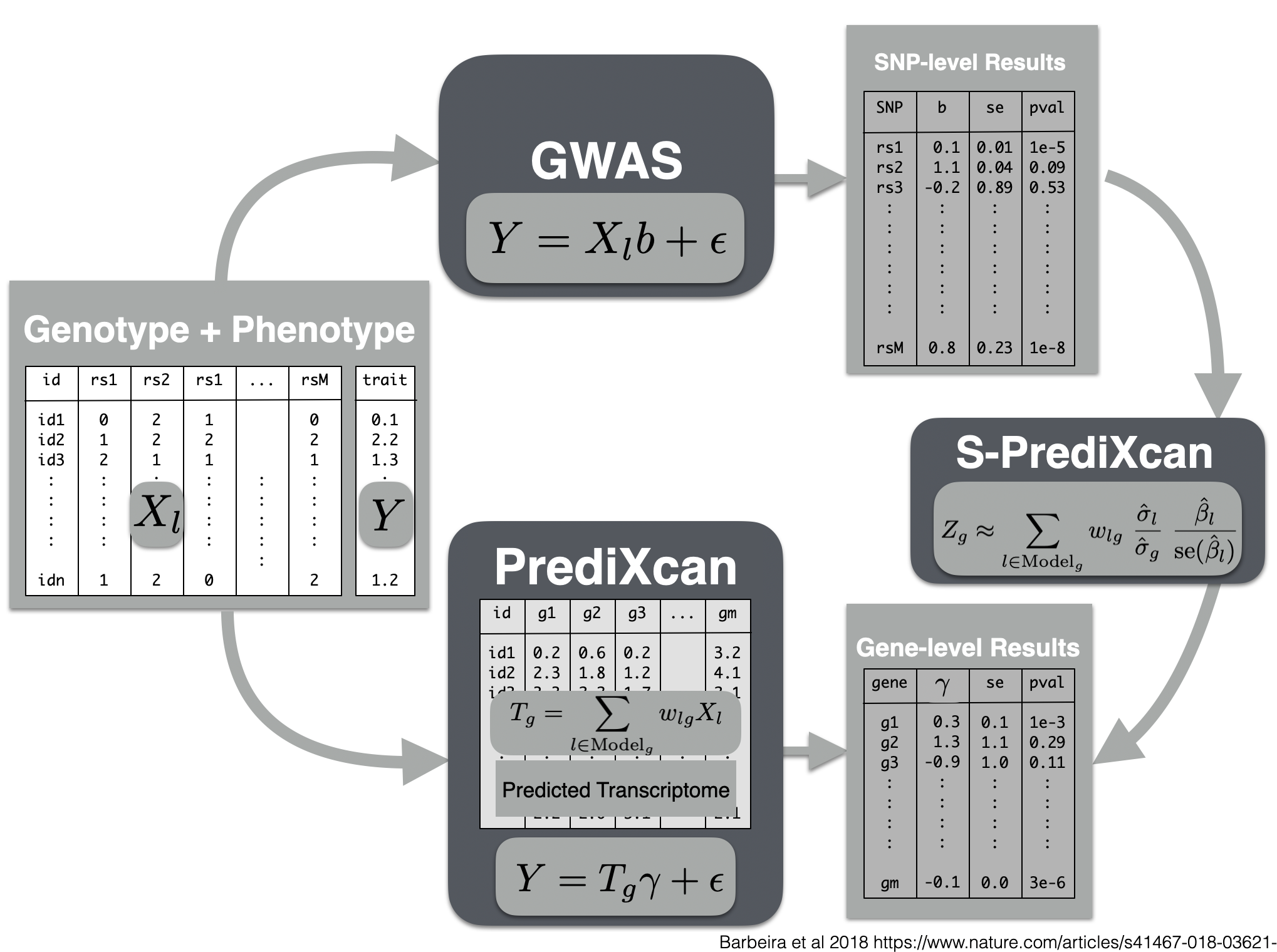
Visual summary of s-predixcan
- harmonized and imputed GWAS result for coronary artery disease is available in $PRE/spredixcan/data/
run s-predixcan
python $METAXCAN/SPrediXcan.py \
--gwas_file $DATA/spredixcan/imputed_CARDIoGRAM_C4D_CAD_ADDITIVE.txt.gz \
--snp_column panel_variant_id --effect_allele_column effect_allele --non_effect_allele_column non_effect_allele --zscore_column zscore \
--model_db_path $MODEL/gtex_v8_mashr/mashr_Whole_Blood.db \
--covariance $MODEL/gtex_v8_mashr/mashr_Whole_Blood.txt.gz \
--keep_non_rsid --additional_output --model_db_snp_key varID \
--throw \
--output_file $RESULTS/spredixcan/eqtl/CARDIoGRAM_C4D_CAD_ADDITIVE__PM__Whole_Blood.csv
plot and interpret s-predixcan results
spredixcan_association = load_spredixcan_association(glue::glue("{RESULTS}/spredixcan/eqtl/CARDIoGRAM_C4D_CAD_ADDITIVE__PM__Whole_Blood.csv"), gencode_df)
dim(spredixcan_association)
spredixcan_association %>% arrange(pvalue) %>% head
spredixcan_association %>% arrange(pvalue) %>% ggplot(aes(pvalue)) + geom_histogram(bins=20)
gg_qqplot(spredixcan_association$pvalue)- SORT1, considered to be a causal gene for LDL cholesterol and as a consequence of coronary artery disease, is not found here. Why? (tissue)
Exercise
run s-predixcan with liver model, do you find SORT1? Is it significant?
compare zscores in liver and whole blood.
run multixcan (optional)
- multixcan aggregates information across multiple tissues to boost the power to detect association. It was developed movivated by the fact that eQTLs are shared across multiple tissues, i.e. many genetic variants that regulate expression are common across tissues.
python $METAXCAN/SMulTiXcan.py \
--models_folder $MODEL/gtex_v8_mashr \
--models_name_pattern "mashr_(.*).db" \
--snp_covariance $MODEL/gtex_v8_expression_mashr_snp_smultixcan_covariance.txt.gz \
--metaxcan_folder $RESULTS/spredixcan/eqtl/ \
--metaxcan_filter "CARDIoGRAM_C4D_CAD_ADDITIVE__PM__(.*).csv" \
--metaxcan_file_name_parse_pattern "(.*)__PM__(.*).csv" \
--gwas_file $DATA/spredixcan/imputed_CARDIoGRAM_C4D_CAD_ADDITIVE.txt.gz \
--snp_column panel_variant_id --effect_allele_column effect_allele --non_effect_allele_column non_effect_allele --zscore_column zscore --keep_non_rsid --model_db_snp_key varID \
--cutoff_condition_number 30 \
--verbosity 7 \
--throw \
--output $RESULTS/smultixcan/eqtl/CARDIoGRAM_C4D_CAD_ADDITIVE_smultixcan.txt
Colocalization methods
- Colocalization methods seek to estimate the probability that the complex trait and expression causal variants are the same. We favor methods that calculate the probability of causality for each trait (posterior inclusion probability), called fine-mapping methods. Here we use torus for fine-mapping and fastENLOC for colocalization.
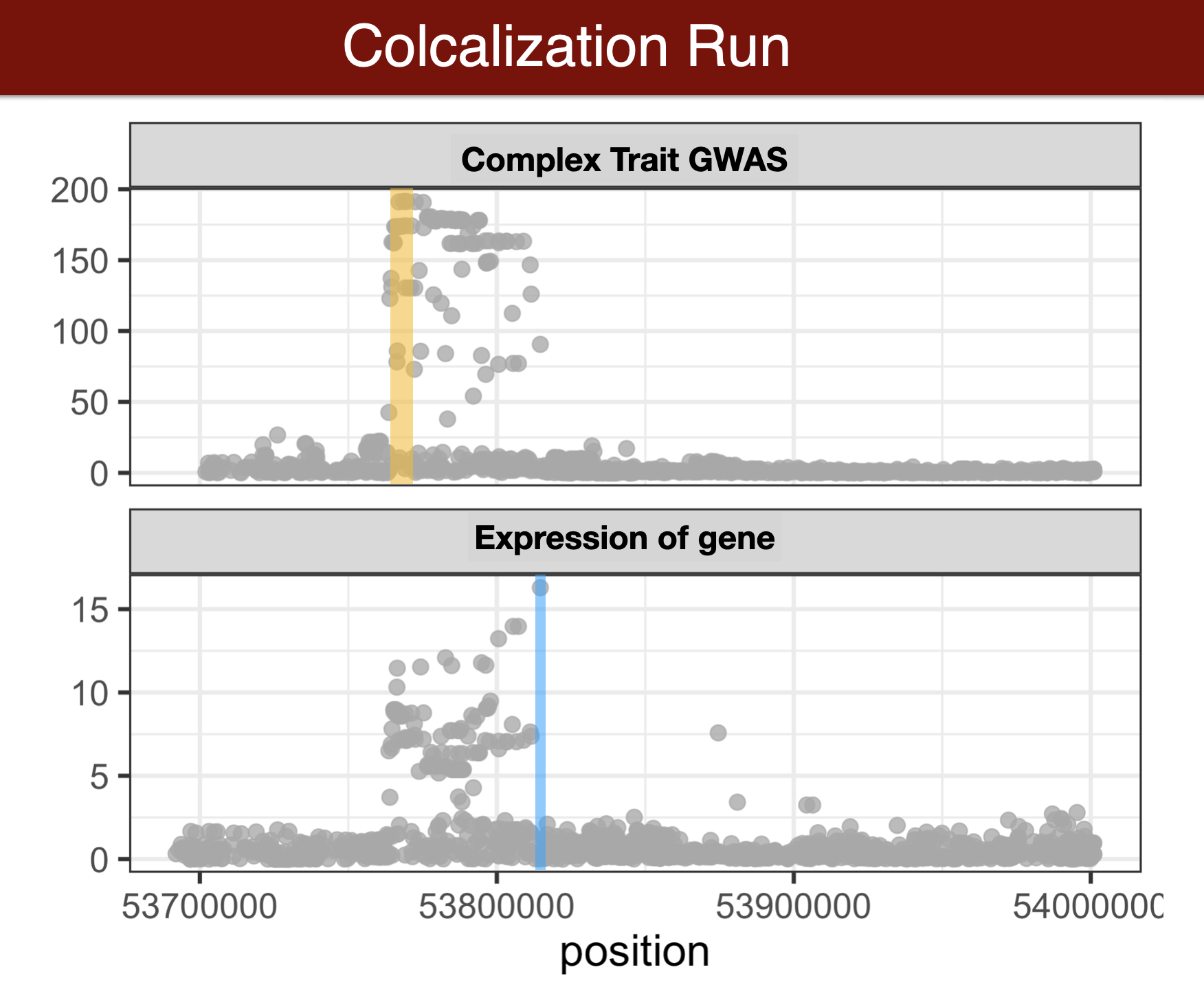
Visual summary of colocalization
GWAS summary statistics to torus format
the following code will format GWAS summary statistics into a format that the fine-mapping method torus can understand.
we precalculated this for you so there is no need to recalculate
## We ran this formatting for you because it takes over 10 minutes.
python $CODE/gwas_to_torus_zscore.py \
-input_gwas $DATA/spredixcan/imputed_CARDIoGRAM_C4D_CAD_ADDITIVE.txt.gz \
-input_ld_regions $DATA/spredixcan/eur_ld_hg38.txt.gz \
-output_fp $DATA/fastenloc/CARDIoGRAM_C4D_CAD_ADDITIVE.zval.gzfine-map GWAS results
We will run torus due to time limitation but ideally we would like to run a method that allows multiple causal variants per locus, such as DAP-G or SusieR.
torus has been precompiled and placed within the PATH
TORUSOFT=torus
$TORUSOFT -d $PRE/data/fastenloc/CARDIoGRAM_C4D_CAD_ADDITIVE.zval.gz --load_zval -dump_pip $PRE/data/fastenloc/CARDIoGRAM_C4D_CAD_ADDITIVE.gwas.pip
cd $PRE/data/fastenloc
gzip CARDIoGRAM_C4D_CAD_ADDITIVE.gwas.pip
cd $PRE We can take a quick look at the z-values and finemapping PIPs:
cd $PRE/data/fastenloc
zless CARDIoGRAM_C4D_CAD_ADDITIVE.zval.gz
zless CARDIoGRAM_C4D_CAD_ADDITIVE.gwas.pip.gzcalculate colocalization with fastENLOC
## check out tutorial https://github.com/xqwen/fastenloc/tree/master/tutorial
eqtl_annotation_gzipped=$PRE/data/fastenloc/FASTENLOC-gtex_v8.eqtl_annot.vcf.gz
gwas_data_gzipped=$PRE/data/fastenloc/CARDIoGRAM_C4D_CAD_ADDITIVE.gwas.pip.gz
TISSUE=Whole_Blood
FASTENLOCSOFT=fastenloc
##FASTENLOCSOFT=/Users/owenmelia/projects/finemapping_bin/src/fastenloc/src/fastenloc
mkdir $RESULTS/fastenloc/
cd $RESULTS/fastenloc/
$FASTENLOCSOFT -eqtl $eqtl_annotation_gzipped -gwas $gwas_data_gzipped -t $TISSUE
#[-total_variants total_snp] [-thread n] [-prefix prefix_name] [-s shrinkage]
analyze results
## optional - compare with s-predixcan results
fastenloc_results = load_fastenloc_coloc_result(glue::glue("{RESULTS}/fastenloc/enloc.sig.out"))
spredixcan_and_fastenloc = inner_join(spredixcan_association, fastenloc_results, by=c('gene'='Signal'))
ggplot(spredixcan_and_fastenloc, aes(RCP, -log10(pvalue))) + geom_point()
## which genes are both colocalized (rcp>0.10) and significantly associated (pvalue<0.05/number of tests)Mendelian randomization methods
run TWMR (for a locus)
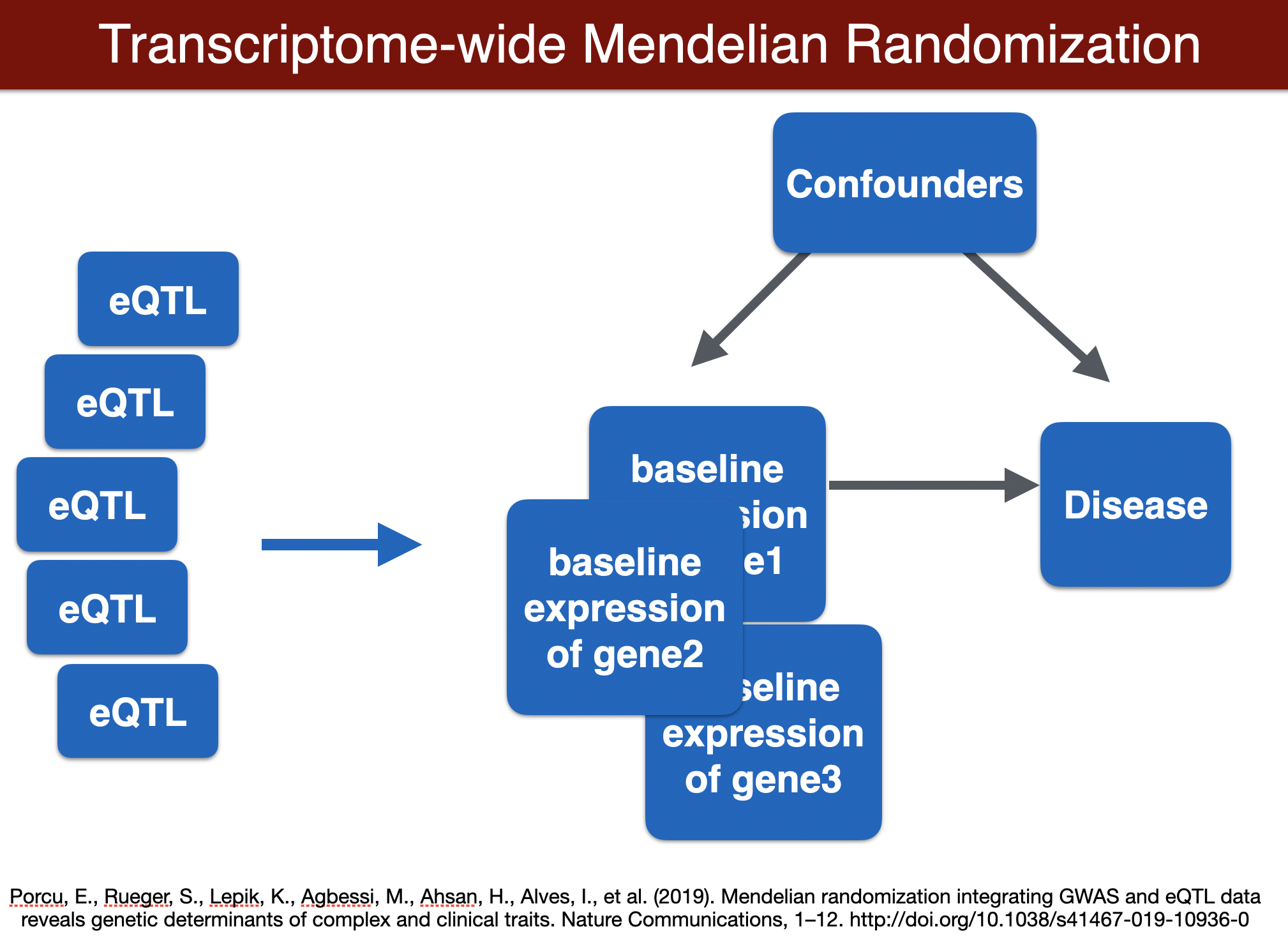
TWMR
# Load the 'analyseMR' function
source(glue::glue("{CODE}/TWMR_script.R"))
# Collect the list of genes available to run
gene_lst <- list.files(TWMR)
gene_lst <- gene_lst[str_detect(gene_lst, "ENS.*")]
gene_lst <- (gsub("\\..*", "", gene_lst) %>% unique)
# Set the gene and run. The function writes output to a file.
for (gene in gene_lst) {
analyseMR(gene, TWMR)
}twmr_results <- load_twmr_results(TWMR, gencode_df)